Delong Zhou, Ph.D.
Bioinformatics Researcher
Exploring RNA Regulation in Disease Through RNA-Seq
Bridging Bioinformatics with Experimental Biology
Bioinformatics Researcher
Exploring RNA Regulation in Disease Through RNA-Seq
Bridging Bioinformatics with Experimental Biology


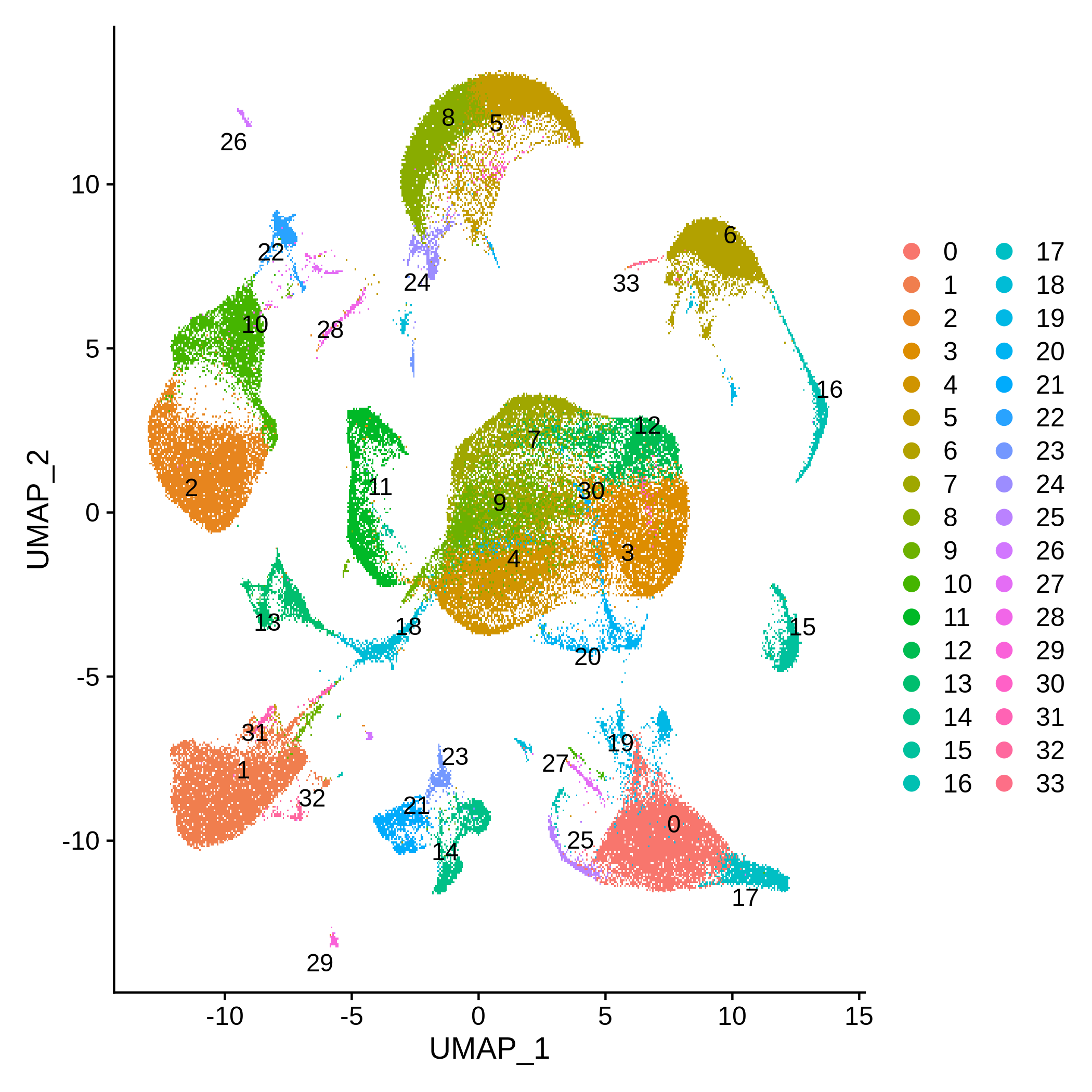
My Current Post-Doc research project at LÉCUYER Lab of RNA Biology at Institut de recherches cliniques de Montréal (IRCM) aims to predict RNA localization signals by training machine learning models on RNA-seq data generated in the lab. By comparing RNA localization signals across different contexts—from cancer- or neurodegenerative patient-derived cell lines to Drosophila cell lines—we aim to determine how RNA localization rules are conserved or divergent and gain deeper insights into their regulation across physiological and pathological conditions.
My first Post-Doc research project under Dr. Bagot at McGill University aims to determine how a depression-impacted brain region responds to psilocybin, a potential cure for people unresopnsive to current treatments. The findings will improve our understanding of how psilocybin works, therefore support the development of psilocybin and other novel depression treatments.
My PhD project supervised by Dr. Michelle Scott and Dr. Sherif Abou Elela at University of Sherbrooke studied how alternative splicing is regulated and how it impacts cell function. Alternative splicing allows one gene to produce multiple proteins of different or sometimes even opposite functions. My work helps to explain how the same splicing regulators can cause opposite outcomes in different tissues. I also developed SAPFIR, a webserver to annotate how changes in splicing lead to changes in protein function to help analyze RNA-seq experiments.
My internship supervised by Dr. Lluis Mir at UMR 8203 Laboratoire de Vectorologie et Thérapeutiques Anti-cancéreuses at CNRS-Institut Gustave-Roussy, Villejuif, France investigated how short bursts of electronic pulses destabilize the cell membranes to introduce chemicals into the cells or to fuse multiple cells. This technique is used to manipulate cytosolic calcium concentration or to fuse cells with different interesting properties to create a hybrid cell line with those properties combined.
In this project lead by Heike Schuler in Dr. Bagot's lab, we challenge the traditional male-centric metrics in depression research by showing that female mice respond differently to social stress. We show that female mice display unique changes in movement patterns rather than typical social avoidance behaviors. We developed a new metric, based on these movement changes, to better study stress in females. This research highlights the importance of tailoring depression studies to account for differences between sexes. I helped to train the machine learning model to track mouse body parts.

In this collaboration between Dr. Scott's lab and Dr. Auger-Messier's lab, we investigated the role of Srsf3 in cardiac function by selectively deleting the gene in heart muscle cells. Our findings demonstrate that loss of Srsf3 disrupts mitochondrial integrity and energy production, leading to cardiomyocyte hypertrophy, impaired contractile function, and early postnatal lethality in mice. These results highlight the critical role of Srsf3 in maintaining mitochondrial homeostasis and cardiac health, providing insights into potential mechanisms underlying mitochondrial dysfunction in heart disease. I analyzed the RNAseq experiment in this study.